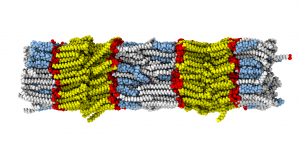
Model membranes and their interaction with small molecules and peptides
We are interested in the structure and dynamics of lipid bilayers (as models for cell membranes), their domain structure and the interaction of small molecules with these bilayers. To this end, we investigate lipid membranes in the presence of a wide range of very different molecules such sterols (especially cholesterol and its derivatives), neurotransmitters (serotonin, melatonin etc.), ceramides, small peptides (NPY, transmembrane helices), drugs (NSAID, statins, small-molecule kinase inhibitors). Also synthetic lipophilic molecules, such as fluorescent probes or ionic liquids, are studied.
With a broad variety of different NMR techniques and other methods (fluorescence and EPR spectroscopy, DSC) we study the influence of these molecules on the lipid membranes as well as their binding, localization, orientation and the dynamic behavior of these molecules in the membrane.
The information obtained in this research will help to understand the interactions of such molecules with cell membranes and help to improve our understanding of the mode the action of medically relevant molecules.
Key Papers:
- Engberg, O., Scheidt, H.A., Nyholm, T.K.M., Slotte, P., Huster, D. Membrane localization and lipid interactions of common lipid-conjugated fluorescence probes. Langmuir 35 (2019) 11902-11911.
- Galiullina LF, Scheidt HA, Huster D, Aganov A, Klochkov V. Interaction of statins with phospholipid bilayers studied by solid-state NMR spectroscopy Biochim Biophys Acta 1861(2019):584-593
- Frits Kamp, Holger A. Scheidt, Edith Winkler, Gabriele Basset, Hannes Heinel, James M. Hutchison, Loren M. LaPointe, Charles R. Sanders, Harald Steiner, Daniel Huster „Binding of bexarotene to the amyloid precursor protein transmembrane domain in liposomes alters its α-helical conformation but inhibits -secretase non-selectively” ACS Chem. Neurosci. 9 (2018) 1702-1713.
- Vogel, A., Scheidt, H.A., Baek, D.J., Bittman, R., Huster, D. Structure and dynamics of the aliphatic cholesterol side chain in membranes as studied by 2H NMR spectroscopy and molecular dynamics simulation. Phys. Chem. Chem. Phys. 18 (2016) 3730-3738.
- Scheidt, H.A., Meyer, T., Nikolaus, J., Baek D.J., Haralampiev, I., Thomas, L., Bittman, R., Müller, P., Herrmann, A., Huster, D. “Cholesterol’s aliphatic side chain modulates membrane properties”. Angew. Chemie Int. Ed. 52 (2013) 12848-12851.
- Bosse, M., Sibold, J., Scheidt, H.A., Patalag, L.J., Kettelhoit, K., Ries, A., Werz, D.B., Steinem, C., Huster, D. Shiga toxin binding alters packing and the domain structure of Gb3-containg membranes: A solid-state NMR study. Phys. Chem. Chem. Phys. 21 (2019) 15630-15638
- Heinrich, F., Salyapongse, A., Kumagai, A., Dupuy, F. G., Shukla, K., Penk, A., Huster, D., Ernst, R. K., Pavlova, A., Gumbart, J. C., Deslouches, B., Di, Y. P., & Tristram-Nagle, S. Synergistic biophysical techniques reveal structural mechanisms of engineered cationic antimicrobial peptides in lipid model membranes. Chemistry 26 (2020) 6247–6256
- Luck, M., Fischer, M., Werle, M., Scheidt, H.A., Müller, P. Impact of selected small-molecule kinase inhibitors on lipid membranes. Pharmaceuticals 14 (2021) 746.
- Haralampiev, I., Alonso, D.J., Luck, M., Fischer, M., Abel, T., Huster, D., Di Lella, S., Scheidt, H.A., Müller, P. Interaction of the small-molecule kinase inhibitors tofacitinib and lapatinib with membranes. Bioch. Biohys. Acta – Biomembranes 1862 (2020) 183414.
- Kaur, N., Fischer, M., Kumar, S., Gahlay, G.K., Scheidt, H.A., Mithu, V.S. Role of cationic head-group in cytotoxicity of ionic liquids: Probing changes in bilayer architecture using solid-state NMR spectroscopy. J. Colloid Interface Sci. 581 (2021) 954-963.
Collaborations:
- Prof. Dr. Andreas Herrmann and Dr. Peter Müller, Humboldt-Universität Berlin, Germany
- Dr. Venus Singh Mithu, Guru Nanak Dev University, Amritsar, India
- Prof. Dr. Vladimir Klochkov, Kazan Federal University, Russia
- Prof Dr. Sudipta Maiti, Tata Institute of Fundamental Research, Mumbai, India
- Prof. Dr. Dieter Langosch, TU München, Germany
- Prof. Dr. Rainer Böckmann, Friedrich-Alexander-Universität Erlangen, Germany
Dynamics of transmembrane helices that are substrates for intramembrane proteases
Intramembrane proteases are cleavage enzymes, which reside in the cell membrane. In contrast to soluble proteases, the mechanism that governs the recognition of substrates for intramembrane proteases is not understood. The goal in this project is to investigate the molecular basis for the target recognition of intramembrane proteases. We hypothesize that there is connection between substrate dynamics and the recognition by the respective intramembrane protease. Therefore, we investigate the local dynamics of known substrates of the intramembrane protease γ-secretase and introduce mutations that change the dynamics of these helices.
For this purpose, we prepare the transmembrane domain of the substrate in different lipid-bilayer systems and measure the local flexibility of the transmembrane domain at certain points. This is realized by various kinds of solid-state NMR spectroscopy methods. With these methods, we investigate the influence of mutations in the transmembrane domain of substrates and the influence of the lipid environment on the dynamic behavior of the substrates. Close collaboration with other colleagues enables us to correlate these results with molecular dynamics simulations and changes in the cleavage behavior of the intramembrane protease. This will provide a deeper insight in the rules which determine these processes on a molecular level.
Key Papers:
- Penk, A., Müller, M., Scheidt, H.A., Langosch, D., Huster D. Structure and Dynamics of the Lipid Modifications of a Transmembrane α-Helical Peptide Determined by 2H Solid-State NMR Spectroscopy. Biochim. Biophys. Acta (Biomembranes) 1808 (2011) 784-791
- B., Langosch, D., Scharnagl, C., Muhle-Goll, C., Steiner, H. Modulating hinge flexibility in the APP transmembrane domain alters γ-secretase cleavage. Biophys. J. 116 (2019) 2103-2120.
- Götz, A., Kamp, F., Mylonas, N., Högel, P., Silber, M., Heinel, H., Menig, S., Vogel, A., Feyrer, H., Huster, D., Luy, B., Langosch, D., Scharnagl, C., Muhle-Goll, C., Steiner, H. Modulating hinge flexibility in the APP transmembrane domain alters γ-secretase cleavage. Biophys. J. 116 (2019) 2103-2120.
Collaborations:
- Dr. Christina Scharnagl, Technische Universität München
- Prof. Dr. Harald Steiner, Ludwig-Maximilians-Universität München
- Prof. Dr. Dieter Langosch, Technische Universität München
A new model for the interpretation of transmembrane helix dynamics

In our research, we often work with single membrane-spanning helices. To investigate their dynamics DIPSHIFT order parameters are measured routinely, but the interpretation remains difficult. To aid the interpretation of the NMR results, we are currently establishing a new model derived from the GALA (geometric analysis of labeled alanines) model from literature. The experimental input of this model are 2H NMR order parameters of labeled alanines in the peptide that are measured in oriented samples. The interpretation of these order parameters is achieved by a geometrical model, in which the orientation of the helix is described by a tilt and an azimuth angle and, depending on the exact implementation, sometimes by additional distribution widths of these angles.
In our work we are now aiming to extend this model to also work for DIPSHIFT order parameters measured for unoriented samples under magic angle spinning. The main advantage of this approach is that the labeled positions are not restricted to alanine, which is why it works for a much wider range of peptide sequences, and the complicated preparation of oriented samples can be avoided. Furthermore, several order parameters can be measured with one sample, thereby significantly reducing the number of required peptides with different labeling schemes.
Publications:
in preparation
Collaborations:
Dr. Christina Scharnagl, Technische Universität München
Structure of stratum corneum lipid model membranes
The stratum corneum is the outermost layer of the skin. It protects the organism against pathogens and prevents dehydration. It can be imagined as a brick and mortar model of Corneocytes embedded in a highly organized lipid matrix. For our studies, the intercellular lipid matrix is of particular interest, consisting of ceramides (often with very long hydrocarbon chains), free fatty acids, and cholesterol at an about equimolar ratio. Despite the importance of this barrier, its basic structure and the influence of ceramide classes and chain lengths on its properties are still poorly understood. Therefore, we investigate a number of model systems, where we exchange different ceramides species or vary their chain lengths to obtain information on the basic physical properties of the model membranes. We employ solid-state 13C and 2H NMR spectroscopy and closely collaborate with groups using neutron scattering, X-ray diffraction and synthesizing the molecules with various labeling schemes.
Key papers:
- Kováčik, A., Vogel, A,, Adler J,, Pullmannová, P,, Vávrová, K., Huster, D.
Probing the role of ceramide hydroxylation in skin barrier lipid models by 2H solid-state NMR spectroscopy and X-ray powder diffraction.
BBA – Biomembranes 1860 (2018) 1162–1170 - Schroeter, A., Stahlberg, S., Školová, B., Sonnenberger, S., Eichner, A., Huster, D., Vávrová, K., Hauß, T., Dobner, B., Neubert, R.H.H., Vogel, A.
Phase separation in stratum corneum lipid model membranes based on ceramide [NP]: neutron diffraction and solid-state NMR.
Soft Mat. 13 (2017) 2107-2119. - Eichner, A., Stahlberg, S., Sonnenberger, S., Lange, S., Dobner, B., Ostermann, A., Schrader, T.E., Hauß, T., Schröter, A., Huster, D., Neubert, R.H.H.
Influence of the penetration enhancer isopropyl myristate on stratum corneum lipid model membranes revealed by neutron diffraction and 2H NMR experiments.
Biochim. Biophys. Acta 1859 (2017) 745-755. - Oliveira, J.S.L., Lange, S., Dobner, B., Brezesinski, G.
The effect of non-deuterated and deuterated isopropyl myristate on the thermodynamical and structural behavior of a 2D stratum corneum model with ceramide [AP].
Chem. Phys. Lipids 204 (2017) 1-9. - Stahlberg, S., Lange, S., Dobner, B., Huster, D.
Probing the role of ceramide headgroup polarity in short-chain model skin barrier lipid mixtures by 2H solid-state NMR spectroscopy.
Langmuir 32 (2016) 2023-2031. - Stahlberg, S., Školová. B., Madhu, P.K., Vogel, A., Vávrová, K., Huster, D.
Probing the role of ceramide acyl chain length and sphingosine unsaturation in model skin barrier lipid mixtures by 2H solid-state NMR spectroscopy.
Langmuir 31 (2015) 4906-4915. - Školová, B., Hudská, K., Pullmannová, P., Kováčik, A., Palát, K., Roh, J., Fleddermann, J., Estrela-Lopis, I., Vávrová, K.
Different phase behavior and packing of ceramides with long (C16) and very long (C24) acyls in model membranes: infrared spectroscopy using deuterated lipids.
J. Phys. Chem.B, 118 (2014) 10460-10470.
Collaborations:
Prof. Pharm. Dr. Kateřina Vávrová, Charles University, Hradec Kralove, Czech Republic