Proteinformatics Workgroup








Research
Proteinformatics
Analysis, prediction and visualization of molecular complexes
Our goal is to improve the understanding of proteins and their functions. Therefore, we use knowledge based predictions of secondary and tertiary structure interactions of helical membrane proteins.
Furthermore, we increase the explanatory power of protein models by fragment based modelling of proteins and protein complexes using Cryo-Electron Microscopy (EM) data as constraints.
In order to share our findings with the research community we are developing web applications for analysis, prediction and visualization of proteins and their complexes.
Structural dynamics of G protein coupled receptors (GPCRs)
Principal Investigator
Prof. Dr. Peter W. Hildebrand

Härtelstr. 16-18, 04107 Leipzig
Room 251
Phone +49(0)341 97-15712
Email
Consultation by email appointment
Secretariat
Jessika Schlögl
Room 255.1
Phone +49(0)341 97-15780
Email
Office hours:
Mo – Fri 9:00 – 13:00
Team
Batebi, Hossein, Dr. rer. nat., guest scientist
Geraili Daronkola, Hosein, Dr. rer. nat., PostDoc
Pankonin, Maik, doctoral student
Seufert, Florian, doctoral student
Staritzbichler, René, Dr. rer. nat., guest scientist
Vogel, Alexander, Dr. rer. nat., research associate, IT support
Salomon, Maximilian, PhD student
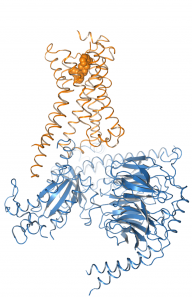
We work on the elucidation of the conformational space of GPCRs and its modulation by extracellular ligands and intracellular effector proteins by means of classical and umbrella sampling molecular dynamics simulations to address the issues of signaling specificity and biased signaling.
For this reason, we are modelling receptor G protein or arrestin interactions computationally to elucidate the mechanism of signal transduction.
Publications
Journal Articles
78. Batebi, H., Pérez-Hernández, G., Rahman, S. N., Lan, B., Kamprad, A., Shi, M., Speck, D., Tiemann, J. K. S., Guixà-González, R., Reinhardt, F., Stadler, P. F., Papasergi-Scott, M. M., Skiniotis, G., Scheerer, P., Kobilka, B. K., Mathiesen, J. M., Liu, X., & Hildebrand, P. W. Mechanistic insights into G-protein coupling with an agonist-bound G-protein-coupled receptor. Nature structural & molecular biology (2024). [DOI; PubMed]
77. Philipp, M., Moth, C.W., Ristic, N., Tiemann, J.K. S., Seufert, F., Panfilova, A., Meiler, J., Hildebrand, P.W., Stein, A., Wiegreffe, D., Staritzbichler, R. MutationExplorer: a webserver for mutation of proteins and 3D visualization of energetic impacts. Nucleic Acids Res (2024). [DOI; PubMed]
76. Papasergi-Scott M.M., Pérez-Hernández G., Batebi H., Gao Y., Eskici G., Seven A.B., Panova O., Hilger D., Casiraghi M., He F., Maul L., Gmeiner P., Kobilka B.K., Hildebrand P.W., Skiniotis G. Time-resolved cryo-EM of G-protein activation by a GPCR. Nature Mar (2024) 13. [DOI; PubMed]
75. Vieira Contreras, F., Auger, G. M., Müller, L., Richter, V., Huetteroth, W., Seufert, F., Hildebrand, P. W., Scholz, N., Thum, A. S., Ljaschenko, D., Blanco-Redondo, B., Langenhan, T. The adhesion G-protein-coupled receptor mayo/CG11318 controls midgut development in Drosophila. Cell reports. 43(1) (2024) 113640. [DOI; PubMed]
74. Seufert, F., Chung, Y. K., Hildebrand, P. W., Langenhan, T. 7TM domain structures of adhesion GPCRs: what’s new and what’s missing?. Trends in Biochemical Sciences (2023) 14. [DOI; PubMed]
73. Ebstein, F., Küry, S., Most, V., Rosenfelt, C., Scott-Boyer, M. P., van Woerden, G. M., Besnard, T., Papendorf, J. J., Studencka-Turski, M., Wang, T., Hsieh, T. C., Golnik, R., Baldridge, D., Forster, C., de Konink, C., Teurlings, S. M. W., Vignard, V., van Jaarsveld, R. H., Ades, L., Cogné, B., Mignot, C., Deb, W., Jongmans, M.C.J., Cole, F.S., van den Boogaard, M.H., Wambach, J.A., Wegner, D.J., Yang, S., Hannig, V., Brault, J.A., Zadeh, N., Bennetts, B., Keren, B., Gélineau, A.C., Powis, Z., Towne, M., Bachman, K., Seeley, A., Beck, A.E., Morrison, J., Westman, R., Averill, K., Brunet, T., Haasters, J., Carter, M.T., Osmond, M., Wheeler, P.G., Forzano, F., Mohammed, S., Trakadis, Y., Accogli, A., Harrison, R., Guo, Y., Hakonarson, H., Rondeau, S., Baujat, G., Barcia, G., Feichtinger, R.G., Mayr, J.A., Preisel, M., Laumonnier, F., Kallinich, T., Knaus, A., Isidor, B., Krawitz, P., Völker, U., Hammer, E., Droit, A., Eichler, E.E., Elgersma, Y., Hildebrand, P.W., Bolduc, F., Krüger, E., Bézieau, S. PSMC3 proteasome subunit variants are associated with neurodevelopmental delay and type I interferon production. Science translational medicine 15(698) (2023) eabo3189. [DOI; PubMed]
72. Smith, A. A., Pacull, E. M., Stecher, S., Hildebrand, P. W., Vogel, A., & Huster, D. Analysis of the Dynamics of the Human Growth Hormone Secretagogue Receptor Reveals Insights into the Energy Landscape of the Molecule. Angew Chem Int Ed Engl 19 (2023) e202302003. [DOI; PubMed]
71. Hunold, P., Fischer, M., Olthoff, C., Hildebrand, P.W., Kaiser, T., Staritzbichler, R. Detecting Pre-Analytically Delayed Blood Samples for Laboratory Diagnostics Using Raman Spectroscopy. Int. J. Mol. Sci. 24 (2023) 7853. [DOI; PubMed]
70. Heng, J., Hu, Y., Pérez-Hernández, G., Inoue, A., Zhao, J., Ma, X., Sun, X., Kawakami, K., Ikuta, T., Ding, J., Yang, Y., Zhang, L., Peng, S., Niu, X., Li, H., Guixà-González, R., Jin, C., Hildebrand, P. W., Chen, C., Kobilka, B. K. Function and dynamics of the intrinsically disordered carboxyl terminus of β2 adrenergic receptor. Nature communications 14(1) (2023) 2005. [DOI; PubMed]
69. Staritzbichler, R., Ristic, N., Stapke, T., Hildebrand, P. W. SmoothT-a server constructing low-energy pathways from conformational ensembles for interactive visualization and enhanced sampling. Bioinformatics (Oxford, England) 39(4) (2023) btad176. [DOI; PubMed]
68. Papasergi-Scott, M. M., Pérez-Hernández, G., Batebi, H., Gao, Y., Eskici, G., Seven, A. B., Panova, O., Hilger, D., Casiraghi, M., He, F., Maul, L., Gmeiner, P., Kobilka, B. K., Hildebrand, P. W., Skiniotis, G. Time-resolved cryo-EM of G protein activation by a GPCR. bioRxiv : the preprint server for biology (2023) 2023.03.20.533387. [DOI; PubMed]
67. Sala, D., Batebi, H., Ledwitch, K., Hildebrand, P.W., Meiler, J. Targeting in silico GPCR conformations with ultra-large library screening for hit discovery. Trends in Pharmacological Sciences 44 (2023) 150-161. [DOI; PubMed]
66. Sala, D., Hildebrand, P.W., Meiler, J. Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties. Frontiers in molecular biosciences 10 (2023) 1121962. [DOI; PubMed]
65. Schmidt, P., Vogel, A., Schwarze, B., Seufert, F., Licha, K., Wycisk, V., Kilian, W., Hildebrand, P.W., Mitschang, L. Towards Probing Conformational States of Y2 Receptor Using Hyperpolarized 129Xe NMR. Molecules 28 (2023) 1424. [DOI; PubMed]
64. Canzler, S., Fischer, M., Ulbricht, D., Ristic, N., Hildebrand, P.W., Staritzbichler, R. ProteinPrompt: a webserver for predicting protein-protein interactions. Bioinformatics Advances 2 (2022) vbac059. [DOI; PubMed]
63. Kampfrath, M., Staritzbichler, R., Pérez Hernández, G., Rose, A.S., Tiemann, J.K.S., Scheuermann, G., Wiegreffe, D., Hildebrand, P.W. MDsrv: visual sharing and analysis of molecular dynamics simulations. Nucleic Acids Res. 50 (2022) gkac398. [DOI; PubMed]
62. Staritzbichler, R., Yaklich, E., Sarti, E., Ristic, N., Hildebrand, P.W., Forrest, L.R. AlignMe: an update of the web server for alignment of membrane protein sequences. Nucleic Acids Res. 50 (2022) gkac391. [DOI; PubMed]
61. Philippe, A., Kleinau, G., Gruner, J.J., Wu, S., Postpieszala, D., Speck, D., Heidecke, H., Dowell, S.J., Riemekasten, G., Hildebrand, P.W., Kamhieh-Milz, J., Catar, R., Szczepek, M., Dragun, D., Scheerer, P. Molecular Effects of Auto-Antibodies on Angiotensin II Type 1 Receptor Signaling and Cell Proliferation. Int J Mol Sci 23 (2022) 3984. [DOI; PubMed]
60. Smith, A.A., Vogel, A., Engberg, O., Hildebrand, P.W., Huster, D. A method to construct the dynamic landscape of a bio-membrane with experiment and simulation. Nat. Commun. 13 (2022) 108. [DOI; PubMed]
59. Heyder, N.A., Kleinau, G., Speck, D., Schmidt, A., Paisdzior, S., Szczepek, M., Bauer, B., Koch, A., Gallandi, M., Kwiatkowski, D., Bürger, J., Mielke, T., Beck-Sickinger, A., Hildebrand, P., Spahn, C.M.T., Hilger, D., Schacherl, M., Biebermann, H., Hilal, T., Kühnen, P., Kobilka, B.K., Scheerer, P. Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-α-MSH and setmelanotide. Cell Res. 31 (2021) 1176-1189. [DOI; PubMed]
58. Staritzbichler, R., Hunold, P., Estrela-Lopis, I., Hildebrand, P.W., Isermann, B., Kaiser, T. Raman spectroscopy on blood serum samples of patients with end-stage liver disease. PLoS One 16 (2021) e0256045. [DOI; PubMed]
57. Staritzbichler, R., Ristic, N., Goede, A., Preissner, R., Hildebrand, P.W., Voronoia 4-ever. Nucleic Acids Res. 49 (2021) W685-W690. [DOI; PubMed]
56. Beliu, G., Altrichter, S., Guixà-González, R., Hemberger, M., Brauer, I., Dahse, A.K., Scholz, N., Wieduwild, R., Kuhlemann, A., Batebi, H., Seufert, F., Pérez-Hernández, G., Hildebrand, P.W., Sauer, M., Langenhan, T. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell. 81 (2021) 905-921. [DOI; PubMed]
55. Vogel, A., Bosse, M., Gauglitz, M., Wistuba, S., Schmidt, P., Kaiser, A., Vsevolod, G., Beck-Sickinger, A.G., Hildebrand, P.W., Huster, D. The dynamics of the neuropeptide Y receptor type 1 investigated by solid-state NMR and molecular dynamics simulation. Molecules 25 (2020) 5489. [DOI; PubMed]
54. Ma, X., Hu, Y., Batebi, H., Heng, J., Xu, J., Liu, X., Niu, X., Li, H., Hildebrand, P.W., Jin, C., Kobilka, B.K. Analysis of β2AR-Gs and β2AR-Gi complex formation by NMR spectroscopy. Proc. Natl. Acad. Sci. 117 (2020) 23096-23105. [DOI; PubMed]
53. Rodríguez-Espigares, I., Torrens-Fontanals, M., Tiemann, J.K.S., Aranda-García, D., Ramírez-Anguita, J.M., Stepniewski, T.M., Worp, N., Varela-Rial, A., Morales-Pastor, A., Medel-Lacruz, B., Pándy-Szekeres, G., Mayol, E., Giorgino, T., Carlsson, J., Deupi, X., Filipek, S., Filizola, M., Gómez-Tamayo, J.C., Gonzalez, A., Gutiérrez-de-Terán, H., Jiménez-Rosés, M., Jespers, W., Kapla, J., Khelashvili, G., Kolb, P., Latek, D., Marti-Solano, M., Matricon, P., Matsoukas, M.T., Miszta, P., Olivella, M., Perez-Benito, L., Provasi, D., Ríos, S., R. Torrecillas, I., Sallander, J., Sztyler, A., Vasile, S., Weinstein, H., Zachariae, U., Hildebrand, P.W., De Fabritiis, G., Sanz, F., Gloriam, D.E., Cordomi, A., Guixà-González, R., Selent, J. Publisher Correction: GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat Methods 17 (2020) 861-862. [DOI; PubMed]
52. Rodriguez-Espigares, I., Torrens-Fontanals, M., Tiemann, J.K.S., Aranda-Garcia, D., Ramirez-Anguita, J.M., Stepniewski, T.M., Worp, N., Varela-Rial, A., Morales-Pastor A., Medel-Lacruz, B., Pándy-Szekeres, G., Mayol, E., Giorgino, T., Carlsson, J., Deupi, X., Filipek, S., Filizola, M., Gómez-Tamayo, J.C., Gonzalez, A., Gutiérrez-de-Terán, H., Jiménez-Rosés, M., Jespers, W., Kapla, J., Khelashvili, G., Kolb, P., Latek, D., Marti-Solano, M., Matricon, P., Matsoukas, M.T., Miszta, P., Olivella, M., Perez-Benito, L., Provasi, D., Rios, S., Torrecillas, I.R., Sallander, J., Sztyler, A., Vasile, S., Weinstein, H., Zachariae, U., Hildebrand, P.W., de Fabritiis, G., Sanz, F., Gloriam, D.E., Cordomi, A., Guixà-González, R., Selent, J. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat. Methods 17 (2020) 777-787. [DOI; PubMed]
51. Mayol, E., García-Recio, A., Tiemann, J.K.S., Hildebrand, P.W., Guixà-González, R., Olivella, M., Cordomi, A. HomolWat: a web server tool to incorporate „homologous“ water molecules into GPCR structures. Nucleic Acids Res. W1 (2020) W54-W59. [DOI]; PubMed]
50. Hildebrand, P.W., Rose, A.S., Tiemann, J. K. S. Bringing molecular dynamics simulation data into view. Trends Biochem. Sci. 44 (2019) 902-913. [ DOI; PubMed]
49. Liu, X., Xu, X., Hilger, D., Aschauer, P., Tiemann, J. K. S., Du, Y., Liu, H., Hirata, K., Sun, X., Guixà-González, R., Mathiesen, J.M., Hildebrand, P.W., Kobilka, B. K. Structural insights into the process of GPCR-G protein complex formation. Cell 177 (2019) 1243-1251. [ DOI; PubMed]
48. Saleh, N., Kleinau, G., Heyder, N., Clark, T., Hildebrand, P. W., Scheerer, P. Binding, thermodynamics, and selectivity of a non-peptide antagonist to the melanocortin-4 receptor. Front. Pharmacol. 9 (2018) 560. [DOI; PubMed]
47. Tiemann, J.K.S., Rose, A.S., Ismer, J., Darvish, M.D., Hilal, T., Spahn, C.M.T., Hildebrand, P.W. FragFit: a web-application for interactive modeling of protein segments in cryo-EM density maps. Nucleic Acids Research 46 (2018) W310-W314. [DOI; PubMed]
46. Bittner, H., Guixà-González, R., Hildebrand, P.W. Structural Basis for the interaction of the beta-secretase with copper. Biochim. Biophys. Acta 1860 (2018) 1105-1113. [DOI; PubMed]
45. Elgeti, M., Kazmin, R., Rose, A.S., Szczepek, M., Hildebrand, P.W., Bartl, F.J., Scheerer, P, Hofmann, K.P. The arrestin-1 finger Loop interacts with two distinct conformations of active rhodopsin. J. biol. Chem. 293 (2018) 4403-4410. [DOI; PubMed]
44. Saleh, N., Hucke, O., Kramer, G., Schmidt, E., Montel, F., Lipinski, R., Ferger, B., Clark, T., Hildebrand, P.W., Tautermann, C.S. Multiple binding sites contribute to the mechanism of mixed agonistic and positive allosteric modulators of the cannabinoid CB1 receptor. Angewandte Chemie Int. Ed. 57 (2018) 2580-2585. [DOI; PubMed]
43. Tiemann J, Guixà-González, R, Hildebrand PW, Rose A. MDsrv: viewing and sharing dynamics simulations on the web. Nature Methods 14 (2017) 1123-1124. [DOI; PubMed]
42. Ismer J, Rose AS, Tiemann JKS, Hildebrand, PW. A Fragment based method for modeling of Protein segments into cryo-EM density maps. BMC Bioinformatics 18 (2017) 475 [DOI; PubMed]
41. Worth CL, Kreuchwig F, Tiemann JKS, Kreuchwig A, Ritschel M, Kleinau G, Hildebrand PW, Krause G. GPCR-SSFE 2.0 – a fragment-based molecular modeling web tool for Class A G-Protein coupled receptors. Nucleic Acids Resarch 45 (2017) W408-W415. [DOI; PubMed]
40. Guixà-González R, Albasanz JL, Rodriguez-Espigares I, Pastor M, Sanz F, Martí-Solano M, Manna5 M, Martinez-Seara H, Hildebrand PW, Martín M and Selent J: Membrane cholesterol access into a G-protein-coupled receptor. Nature Commun. 8 (2017) 14505. [PubMed; DOI]
39. Ismer J, Rose AS, Tiemann JK, Goede A, Preissner R, Hildebrand PW.: SL2: an interactive webtool for modeling of missing segments in proteins. Nucleic Acids Res, W1 (2017) W390-394. [PubMed; DOI]
38. Sprink T., Ramrath D, Yamamoto H., Yamamoto K., Loerke J., Ismer J., Hildebrand P.W., Scheerer P., Bürger J., Mielke T. and Spahn CMT. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Science Advances. 3 (2016) e1501502. [PubMed; DOI]
37. Rose A., Zacchariae U., Grubmüller H., Hofmann K.P., Scheerer P. and Hildebrand P.W. Role of Structural Dynamics at the Receptor G Protein Interface for Signal Transduction. PLoS ONE. 11 (2015) e0143399. [PubMed; DOI]
36. Yamamoto H., Collier M., Loerke J., Ismer J., et al., Hildebrand P.W., Scheerer P. and Spahn C.M.T. Molecular architecture of the ribosome-bound Hepatitis C virus internal ribosomal entry site RNA. EMBOJ. 24 (2015) 3042-3058. [PubMed; DOI]
35 Barucker C. et al., Hildebrand P.W. McKinney A. and Multhaup G. Aβ42-oligomer interacting peptide (AIPs) neutralizes toxic amyloid-β42 species and protects synaptic structure and function. Sci Rep. 5 (2015) 15410. [PubMed; DOI]
34. Brehm, A., et al., Hildebrand P.W., Brogan P., Krüger E., Aksentijevich I., Goldbach-Mansky R. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 11 (2015) 4196-4211. [PubMed; DOI]
33. Prade E., Bittner H.J., Sarkar R., Amo J.M.L., Althoff-Ospelt G., Hildebrand P.W. and Reif B. Structural mechanism of the interaction of Alzheimer’s disease Aβ fibrils with the non-steroidal anti-inflammatory drug (NSAID) sulindac sulfide. JBC. 48 (2015) 28737-28745. [PubMed; DOI]
32. Kazmin R, Rose A, Szczepek M, Elgeti M, Ritter E, Piechnick R, Hofmann K.P., Scheerer P and Hildebrand P.W. and Bartl. The activation pathway of human rhodopsin in comparison to bovine rhodopsin. JBC. 33 (2015) 20117-20127. [PubMed; DOI]
31. Rose, A. S. and Hildebrand P.W. NGL Viewer: A web application for molecular visualization. Nucleic Acids Res. 41 (2015) D576-579. [PubMed; DOI]
30. Österreich, F., Bittner, H., Weise, C., Grohmann, L., Janke, L. K., Hildebrand, P.W., Multhaup, G., Munter, L. Impact of amyloid precursor protein hydrophilic transmembrane residues on amyloid-beta generation. Biochemistry. 17 (2015) 2777-2784. [PubMed; DOI]
29. Sommer, M. E., Elgeti, M., Hildebrand, P. W., Szczepek, M., Hofmann, K. P and Scheerer, P. Structure-based biophysical analysis of the interaction of rhodopsin with G Protein and arrestin. Methods Enzymol. 556 (2015) 563-608. [PubMed; DOI]
28. Szczepek, M., Beyrière, F., Hofmann, K. P., Elgeti, M., Kazmin, R., Rose, A., Bartl, F. J., Stetten von, D., Heck, M., Sommer, M. E., Hildebrand, P. W. and Scheerer, P. Crystal structure of a common GPCR binding interface for G protein and arrestin. Nat. Commun. 10 (2014) 4801. [PubMed; DOI]
27. Rose A.S., Elgeti M., Zacchariae U., Grubmüller H., Hofmann K.P., Scheerer P. and Hildebrand P.W. Position of transmembrane helix 6 determines receptor g protein coupling specificity. J Am Chem Soc. 32 (2014) 11244-11247. [PubMed; DOI]
26. Barucker C., Harmeier A., Weiske J., Fauler B., Albring K.F., Prokop S., Hildebrand P.W., Lurz R., Heppner F.L., Huber O., Multhaup G. Nuclear translocation uncovers the amyloid Peptide aβ42 as a regulator of gene transcription. J Biol Chem. 29 (2014) 20182-20191. [PubMed; DOI]
25. Budkevich T.V., Giesebrecht J., Ramrath D., Mielke T., Ismer J., Hildebrand P.W., Tung C.S., Nierhaus K.H., Sanbonmatsu K.Y. and Spahn C.M.T. Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 1 (2014) 121-131. [PubMed; DOI]
24. Munter L.M., Sieg H., Bethge T., Liebsch F., Bierkandt F.S., Schleeger M., Bittner H.J., Heberle J., Jakubowski N. and Hildebrand P.W., Multhaup G. Model peptides uncover the role of the ß-secretase transmembrane sequence in metal-ion mediated oligomerization. J Am Chem Soc. 51 (2013) 19354-19361. [PubMed; DOI]
23. Rose, A., Theune, D., Goede, A. and Hildebrand, P.W. MP:PD—a data base of internal packing densities, internal packing defects and internal waters of helical membrane proteins. Nucleic Acids Res. 42 (2013) D347-D351. [PubMed; DOI]
22. Elgeti M., Rose A.S., Bartl F.J., Hildebrand P.W., Hofmann K.P. and Heck M. Precision vs. flexibility in GPCR signalling. J Am Chem Soc. 33 (2013) 12305-12312. [DOI; PubMed]
21. Ismer, J., Rose, A., Goede, A., Johanna K. S. Tiemann, Rother, K. and Hildebrand, P.W. Voronoia4RNA – A database of atomic packing densities of RNA structures and their complexes. Nucleic Acids Res. 41 (2012) D280-D284. [PubMed; DOI]
20. Kaden,D., Harmeier, A., Weise, C., Munter, L.M., Althoff, V., Rost, B.R., Hildebrand, P.W., Schmitz, D., Schaefer, M., Lurz, R., et al. Novel APP/Aβ mutation K16N produces highly toxic heteromeric Aβ oligomers. EMBO Mol Med. 7 (2012) 647-659. [PubMed; DOI]
19. Piechnick, R., Ritter, E., Hildebrand, P.W., Ernst, O.P., Scheerer, P., Hofmann, K. and Heck, M. The effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc Natl Acad Sci U S A. 109 (2012) 5247-5252. [PubMed]
18. Botev, A., Munter, L.M., Wenzel, R., Richter, L., Althoff, V., Ismer, J., Gerling, U., Weise, C., Koksch, B., Hildebrand, P.W. et al. The Amyloid Precursor Protein C-Terminal Fragment C100 Occurs in Monomeric and Dimeric Stable Conformations and Binds gamma-Secretase Modulators. Biochemistry. 50 (2011) 828-835. [PubMed]
17. Muhs, M., Yamamoto, H., Ismer, J., Takaku, H., Nashimoto, M., Uchiumi, T., Nakashima, N., Mielke, T., Hildebrand, P.W., Nierhaus, K.H. et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 39 (2011) 5264-5275. [PubMed]
16. Ratje, A.H., Loerke, J., Mikolajka, A., Brunner, M., Hildebrand, P.W., Starosta, A.L., Donhofer, A., Connell, S.R., Fucini, P., Mielke, T. et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 468 (2010) 713-716. [PubMed]
15. Richter, L., Munter, L.M., Ness, J., Hildebrand, P.W., Dasari, M., Unterreitmeier, S., Bulic, B., Beyermann, M., Gust, R., Reif, B. et al. (2010) Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc Natl Acad Sci U S A. 107 (2010) 14597-14602. [PubMed]
14. Rose, A., Goede, A. and Hildebrand, P.W. MPlot–a server to analyze and visualize tertiary structure contacts and geometrical features of helical membrane proteins. Nucleic Acids Res. 38 (2010) W602-W608. [PubMed]
13. Munter, L.M., Botev, A., Richter, L., Hildebrand, P.W., Althoff, V., Weise, C., Kaden, D. and Multhaup, G. Aberrant amyloid precursor protein (APP) processing in hereditary forms of Alzheimer disease caused by APP familial Alzheimer disease mutations can be rescued by mutations in the APP GxxxG motif. J Biol Chem. 285 (2010) 21636-21643. [PubMed]
12. Hildebrand, P.W., Scheerer, P., Park, J.H., Choe, H.W., Piechnick, R., Ernst, O.P., Hofmann, K.P. and Heck, M. A ligand channel through the G protein coupled receptor opsin. PLoS ONE. 4 (2009) e4382. [PubMed]
11. Scheerer, P., Heck, M., Goede, A., Park, J.H., Choe, H.W., Ernst, O.P., Hofmann, K.P. and Hildebrand, P.W. Structural and kinetic modeling of an activating helix switch in the rhodopsin-transducin interface. Proc Natl Acad Sci U S A. 106 (2009) 10660-10665. [PubMed]
10. Harmeier, A., Wozny, C., Rost, B.R., Munter, L.M., Hua, H., Georgiev, O., Beyermann, M., Hildebrand, P.W., Weise, C., Schaffner, W. et al. Role of amyloid-beta glycine 33 in oligomerization, toxicity, and neuronal plasticity. J Neurosci. 29 (2009) 7582-7590. [PubMed]
9. Rose, A., Lorenzen, S., Goede, A., Gruening, B. and Hildebrand, P.W. RHYTHM–a server to predict the orientation of transmembrane helices in channels and membrane-coils. Nucleic Acids Res. 37 (2009) W575-580. [PubMed]
8. Hildebrand, P.W., Goede, A., Bauer, R.A., Gruening, B., Ismer, J., Michalsky, E. and Preissner, R. SuperLooper–a prediction server for the modeling of loops in globular and membrane proteins. Nucleic Acids Res. 37 (2009) W571-574. [PubMed]
7. Hofmann, K.P., Scheerer, P., Hildebrand, P.W., Choe, H.W., Park, J.H., Heck, M. and Ernst, O.P. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 34 (2009) 540-552. [PubMed]
6. Rother, K., Hildebrand, P.W., Goede, A., Gruening, B. and Preissner, R. Voronoia: analyzing packing in protein structures. Nucleic Acids Res. 37 (2009) D393-395. [PubMed]
5. Scheerer, P., Park, J.H., Hildebrand, P.W., Kim, Y.J., Krauss, N., Choe, H.W., Hofmann, K.P. and Ernst, O.P. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 455 (2008) 497-502. [PubMed]
4. Hildebrand, P.W., Gunther, S., Goede, A., Forrest, L., Frommel, C. and Preissner, R. Hydrogen-bonding and packing features of membrane proteins: functional implications. Biophys J. 94 (2008) 1945-1953. [PubMed]
3. Hildebrand, P.W., Lorenzen, S., Goede, A. and Preissner, R. Analysis and prediction of helix-helix interactions in membrane channels and transporters. Proteins. 64 (2006) 253-262. [PubMed]
2. Hildebrand, P.W., Rother, K., Goede, A., Preissner, R. and Frommel, C. Molecular packing and packing defects in helical membrane proteins. Biophys J. 88 (2005) 1970-1977. [PubMed]
1. Hildebrand, P.W., Preissner, R. and Frömmel, C. Structural features of transmembrane helices. FEBS Lett. 559 (2004) 145-151. [PubMed]